| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2009-04-06 16:20:42 UTC |
|---|
| Update Date | 2022-03-07 02:51:22 UTC |
|---|
| HMDB ID | HMDB0012188 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | all-trans-Hexaprenyl diphosphate |
|---|
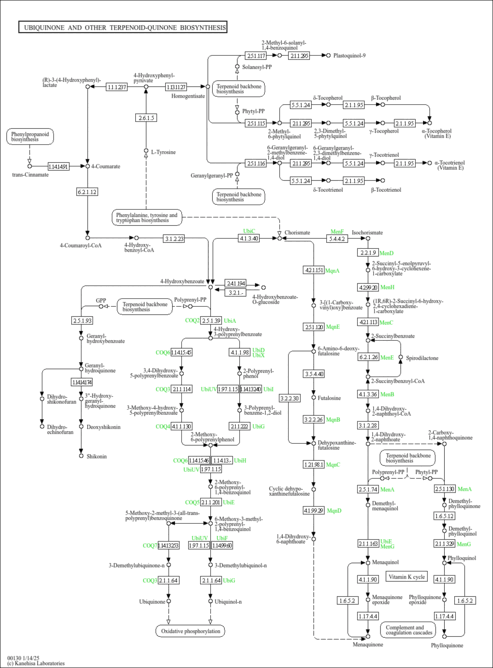
| Description | all-trans-Hexaprenyl diphosphate is the final product of the hexaprenyl diphosphate biosynthesis pathway. In this pathway, multiple units of isopentenyl diphosphate (IPP) undergo a series of polymerizations to form various polyisoprenoids. There are two different pathways for the biosynthesis of IPP. Bacteria that possess ubiquinone generally use the methylerythritol phosphate pathway (MEP), while the eukaryotic microorganisms use the mevalonate pathway. However, exceptions exist. For example, some eukaryotic microbes, like the green algae and the malarial parasite Plasmodium falciparum, appear to utilize the MEP pathway, and some bacteria utilize the mevalonate pathway (Eisenreich01, Eisenreich04). In Saccharomyces cerevisiae S288C, the initial addition of two isoprenyl units to form (E, E)-farnesyl diphosphate is catalyzed by geranyltransferase / dimethylallyltransferase, encoded by FPP1. An additional unit is added by farnesyltranstransferase (encoded by BTS1), resulting in the formation of all-trans-geranyl-geranyl diphosphate. The last enzyme in this pathway is hexaprenyl diphosphate synthase (encoded by COQ1), which adds additional isoprenoid units to a maximal length unique to the organism. In the case of Saccharomyces cerevisiae S288C, it is 6 units. Polyprenyl diphosphate synthase enzymes, such as hexaprenyl diphosphate synthase, are responsible for determining the final length of the tail. When yeast COQ1 mutants are complemented with homologs from other organisms, ubiquinone biosynthesis is restored, but the tail length of the quinone depends on the source of the enzyme. |
|---|
| Structure | CC(C)=CCC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\COP(O)(=O)OP(O)(O)=O InChI=1S/C30H52O7P2/c1-25(2)13-8-14-26(3)15-9-16-27(4)17-10-18-28(5)19-11-20-29(6)21-12-22-30(7)23-24-36-39(34,35)37-38(31,32)33/h13,15,17,19,21,23H,8-12,14,16,18,20,22,24H2,1-7H3,(H,34,35)(H2,31,32,33)/b26-15+,27-17+,28-19+,29-21+,30-23+ |
|---|
| Synonyms | | Value | Source |
|---|
| (2E,6E,10E,14E,18E)-3,7,11,15,19,23-Hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl trihydrogen diphosphate | ChEBI | | (2E,6E,10E,14E,18E)-3,7,11,15,19,23-Hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl trihydrogen diphosphoric acid | Generator | | all-trans-Hexaprenyl diphosphoric acid | Generator | | (E)-Hexaprenyl diphosphate | HMDB |
|
|---|
| Chemical Formula | C30H52O7P2 |
|---|
| Average Molecular Weight | 586.6772 |
|---|
| Monoisotopic Molecular Weight | 586.318827042 |
|---|
| IUPAC Name | [({[(2E,6E,10E,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy}(hydroxy)phosphoryl)oxy]phosphonic acid |
|---|
| Traditional Name | {[(2E,6E,10E,14E,18E)-3,7,11,15,19,23-hexamethyltetracosa-2,6,10,14,18,22-hexaen-1-yl]oxy(hydroxy)phosphoryl}oxyphosphonic acid |
|---|
| CAS Registry Number | 207513-95-9 |
|---|
| SMILES | CC(C)=CCC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\CC\C(C)=C\COP(O)(=O)OP(O)(O)=O |
|---|
| InChI Identifier | InChI=1S/C30H52O7P2/c1-25(2)13-8-14-26(3)15-9-16-27(4)17-10-18-28(5)19-11-20-29(6)21-12-22-30(7)23-24-36-39(34,35)37-38(31,32)33/h13,15,17,19,21,23H,8-12,14,16,18,20,22,24H2,1-7H3,(H,34,35)(H2,31,32,33)/b26-15+,27-17+,28-19+,29-21+,30-23+ |
|---|
| InChI Key | NGFSMHKFTZROKJ-MMSZMYIBSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as bactoprenol diphosphates. These are polyprenyl compounds consisting of a diphosphate group substituted by a bactoprenyl moiety. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Lipids and lipid-like molecules |
|---|
| Class | Prenol lipids |
|---|
| Sub Class | Polyprenols |
|---|
| Direct Parent | Bactoprenol diphosphates |
|---|
| Alternative Parents | |
|---|
| Substituents | - Bactoprenol diphosphate
- Polyprenyl diphosphate
- Polyprenyl monophosphate
- Sesterterpenoid
- Organic pyrophosphate
- Isoprenoid phosphate
- Monoalkyl phosphate
- Alkyl phosphate
- Phosphoric acid ester
- Organic phosphoric acid derivative
- Organic oxygen compound
- Organic oxide
- Hydrocarbon derivative
- Organooxygen compound
- Aliphatic acyclic compound
|
|---|
| Molecular Framework | Aliphatic acyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Physiological effect | Not Available |
|---|
| Disposition | |
|---|
| Process | |
|---|
| Role | |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 9.6 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 17.9003 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 2.09 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 82.8 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 3818.6 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 189.3 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 241.4 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 178.4 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 150.9 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 929.9 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 1064.6 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 107.5 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 1768.0 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 891.0 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 671.9 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 748.1 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 416.1 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 281.3 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 492.2 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 9.0 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized| Derivative Name / Structure | SMILES | Kovats RI Value | Column Type | Reference |
|---|
| all-trans-Hexaprenyl diphosphate,1TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O)O | 4023.6 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O)O | 3221.6 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O)O | 4948.9 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O)O[Si](C)(C)C | 4022.8 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O)O[Si](C)(C)C | 3229.9 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O)O[Si](C)(C)C | 4971.3 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O)O[Si](C)(C)C | 3944.5 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O)O[Si](C)(C)C | 3288.5 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O)O[Si](C)(C)C | 4460.4 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O[Si](C)(C)C)O[Si](C)(C)C | 3946.5 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O[Si](C)(C)C)O[Si](C)(C)C | 3266.5 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O[Si](C)(C)C)O[Si](C)(C)C | 4463.6 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,3TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O[Si](C)(C)C)O[Si](C)(C)C | 3873.1 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,3TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O[Si](C)(C)C)O[Si](C)(C)C | 3324.2 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,3TMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C)OP(=O)(O[Si](C)(C)C)O[Si](C)(C)C | 3921.4 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TBDMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C(C)(C)C)OP(=O)(O)O | 4206.3 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TBDMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C(C)(C)C)OP(=O)(O)O | 3363.0 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TBDMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C(C)(C)C)OP(=O)(O)O | 5006.3 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TBDMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O)O[Si](C)(C)C(C)(C)C | 4201.9 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TBDMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O)O[Si](C)(C)C(C)(C)C | 3377.9 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,1TBDMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O)O[Si](C)(C)C(C)(C)C | 5022.1 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TBDMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C(C)(C)C)OP(=O)(O)O[Si](C)(C)C(C)(C)C | 4317.6 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TBDMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C(C)(C)C)OP(=O)(O)O[Si](C)(C)C(C)(C)C | 3568.5 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TBDMS,isomer #1 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O[Si](C)(C)C(C)(C)C)OP(=O)(O)O[Si](C)(C)C(C)(C)C | 4522.3 | Standard polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TBDMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 4320.4 | Semi standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TBDMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 3537.3 | Standard non polar | 33892256 | | all-trans-Hexaprenyl diphosphate,2TBDMS,isomer #2 | CC(C)=CCC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/CC/C(C)=C/COP(=O)(O)OP(=O)(O[Si](C)(C)C(C)(C)C)O[Si](C)(C)C(C)(C)C | 4554.3 | Standard polar | 33892256 |
|
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted GC-MS | Predicted GC-MS Spectrum - all-trans-Hexaprenyl diphosphate GC-MS (Non-derivatized) - 70eV, Positive | splash10-00tb-7466390000-1a091a15f69aa600fd33 | 2017-09-01 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 10V, Positive-QTOF | splash10-0a4r-0202690000-ca7be5f91d120b34e02f | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 20V, Positive-QTOF | splash10-0a4i-2312910000-a18bd263c2252df7e12b | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 40V, Positive-QTOF | splash10-0a59-4439820000-433602f028fdf8312339 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 10V, Negative-QTOF | splash10-000i-0400090000-eba550c0908e0bdcc04f | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 20V, Negative-QTOF | splash10-056r-7900010000-903a330facd80aef97a7 | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 40V, Negative-QTOF | splash10-004i-9000000000-5db7557b14be93fbdd9b | 2016-09-12 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 10V, Positive-QTOF | splash10-000i-0100290000-dc5d848846327789fd85 | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 20V, Positive-QTOF | splash10-0aos-0004930000-e9910289e743569a483c | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 40V, Positive-QTOF | splash10-020a-1925210000-a29abfabad842bde7acf | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 10V, Negative-QTOF | splash10-000i-0000090000-8f206fdd3583884650db | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 20V, Negative-QTOF | splash10-000i-1300190000-fbac398a5a0b0394af5d | 2021-09-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - all-trans-Hexaprenyl diphosphate 40V, Negative-QTOF | splash10-056r-9400000000-c2d5c8e79c928bb87206 | 2021-09-25 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, H2O, predicted) | 2022-08-20 | Wishart Lab | View Spectrum |
|
|---|