| Record Information |
|---|
| Version | 5.0 |
|---|
| Status | Expected but not Quantified |
|---|
| Creation Date | 2008-08-13 17:53:39 UTC |
|---|
| Update Date | 2023-02-21 17:17:22 UTC |
|---|
| HMDB ID | HMDB0006875 |
|---|
| Secondary Accession Numbers | |
|---|
| Metabolite Identification |
|---|
| Common Name | 1-Pyrroline-2-carboxylic acid |
|---|
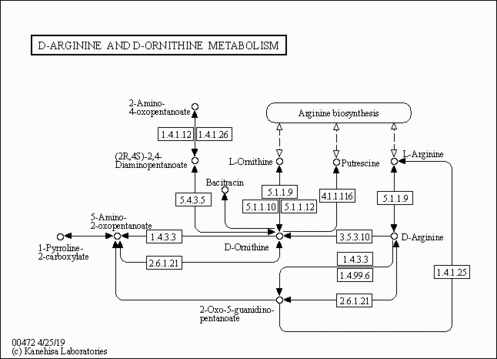
| Description | 1-Pyrroline-2-carboxylic acid, also known as delta1-pyrroline 2-carboxylate, belongs to the class of organic compounds known as pyrrolines. Pyrrolines are compounds containing a pyrroline ring, which is a five-member unsaturated aliphatic ring with one nitrogen atom and four carbon atoms. 1-Pyrroline-2-carboxylic acid is a moderately basic compound (based on its pKa). 1-Pyrroline-2-carboxylic acid exists in all living organisms, ranging from bacteria to humans. 1-pyrroline-2-carboxylic acid can be biosynthesized from D-proline through its interaction with the enzyme D-amino-acid oxidase. The product resulting from formal oxidation of DL-proline by loss of hydrogen from the nitrogen and from the carbon alpha to the carboxylic acid, with the formation of a C=N bond. In humans, 1-pyrroline-2-carboxylic acid is involved in the metabolic disorder called hyperprolinemia type II. |
|---|
| Structure | InChI=1S/C5H7NO2/c7-5(8)4-2-1-3-6-4/h1-3H2,(H,7,8) |
|---|
| Synonyms | | Value | Source |
|---|
| 1-Pyrroline-2-carboxylate | ChEBI | | delta1-Pyrroline 2-carboxylate | ChEBI | | 1-Pyrroline 2-carboxylate | Kegg | | delta1-Pyrroline 2-carboxylic acid | Generator | | Δ1-pyrroline 2-carboxylate | Generator | | Δ1-pyrroline 2-carboxylic acid | Generator | | 1-Pyrroline 2-carboxylic acid | Generator | | 3,4-Dihydro-2H-pyrrole-5-carboxylate | HMDB | | 3,4-Dihydro-2H-pyrrole-5-carboxylic acid | HMDB | | D1-Pyrroline 2-carboxylate | HMDB | | D1-Pyrroline 2-carboxylic acid | HMDB | | delta-1-Pyrroline-2-carboxylate | HMDB | | 1,2-Didehydroproline | HMDB | | 1-Pyrroline-2-carboxylic acid | HMDB | | delta1-Pyrroline-2-carboxylic acid | HMDB | | Δ1-Pyrroline-2-carboxylic acid | HMDB |
|
|---|
| Chemical Formula | C5H7NO2 |
|---|
| Average Molecular Weight | 113.1146 |
|---|
| Monoisotopic Molecular Weight | 113.047678473 |
|---|
| IUPAC Name | 3,4-dihydro-2H-pyrrol-1-ium-5-carboxylate |
|---|
| Traditional Name | 4,5-dihydro-3H-pyrrol-1-ium-2-carboxylate |
|---|
| CAS Registry Number | 2139-03-9 |
|---|
| SMILES | OC(=O)C1=NCCC1 |
|---|
| InChI Identifier | InChI=1S/C5H7NO2/c7-5(8)4-2-1-3-6-4/h1-3H2,(H,7,8) |
|---|
| InChI Key | RHTAIKJZSXNELN-UHFFFAOYSA-N |
|---|
| Chemical Taxonomy |
|---|
| Description | Belongs to the class of organic compounds known as pyrrolines. Pyrrolines are compounds containing a pyrroline ring, which is a five-member unsaturated aliphatic ring with one nitrogen atom and four carbon atoms. |
|---|
| Kingdom | Organic compounds |
|---|
| Super Class | Organoheterocyclic compounds |
|---|
| Class | Pyrrolines |
|---|
| Sub Class | Not Available |
|---|
| Direct Parent | Pyrrolines |
|---|
| Alternative Parents | |
|---|
| Substituents | - Pyrroline
- Ketimine
- Carboxylic acid derivative
- Carboxylic acid
- Monocarboxylic acid or derivatives
- Azacycle
- Organic 1,3-dipolar compound
- Propargyl-type 1,3-dipolar organic compound
- Carbonyl group
- Hydrocarbon derivative
- Organooxygen compound
- Organonitrogen compound
- Organic oxide
- Organopnictogen compound
- Imine
- Organic oxygen compound
- Organic nitrogen compound
- Aliphatic heteromonocyclic compound
|
|---|
| Molecular Framework | Aliphatic heteromonocyclic compounds |
|---|
| External Descriptors | |
|---|
| Ontology |
|---|
| Not Available | Not Available |
|---|
| Physical Properties |
|---|
| State | Solid |
|---|
| Experimental Molecular Properties | | Property | Value | Reference |
|---|
| Melting Point | Not Available | Not Available | | Boiling Point | Not Available | Not Available | | Water Solubility | Not Available | Not Available | | LogP | Not Available | Not Available |
|
|---|
| Experimental Chromatographic Properties | Not Available |
|---|
| Predicted Molecular Properties | |
|---|
| Predicted Chromatographic Properties | Predicted Collision Cross SectionsPredicted Retention Times Underivatized| Chromatographic Method | Retention Time | Reference |
|---|
| Measured using a Waters Acquity ultraperformance liquid chromatography (UPLC) ethylene-bridged hybrid (BEH) C18 column (100 mm × 2.1 mm; 1.7 μmparticle diameter). Predicted by Afia on May 17, 2022. Predicted by Afia on May 17, 2022. | 2.04 minutes | 32390414 | | Predicted by Siyang on May 30, 2022 | 9.3605 minutes | 33406817 | | Predicted by Siyang using ReTip algorithm on June 8, 2022 | 4.87 minutes | 32390414 | | AjsUoB = Accucore 150 Amide HILIC with 10mM Ammonium Formate, 0.1% Formic Acid | 174.5 seconds | 40023050 | | Fem_Long = Waters ACQUITY UPLC HSS T3 C18 with Water:MeOH and 0.1% Formic Acid | 748.1 seconds | 40023050 | | Fem_Lipids = Ascentis Express C18 with (60:40 water:ACN):(90:10 IPA:ACN) and 10mM NH4COOH + 0.1% Formic Acid | 329.9 seconds | 40023050 | | Life_Old = Waters ACQUITY UPLC BEH C18 with Water:(20:80 acetone:ACN) and 0.1% Formic Acid | 98.7 seconds | 40023050 | | Life_New = RP Waters ACQUITY UPLC HSS T3 C18 with Water:(30:70 MeOH:ACN) and 0.1% Formic Acid | 215.5 seconds | 40023050 | | RIKEN = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 86.5 seconds | 40023050 | | Eawag_XBridgeC18 = XBridge C18 3.5u 2.1x50 mm with Water:MeOH and 0.1% Formic Acid | 276.6 seconds | 40023050 | | BfG_NTS_RP1 =Agilent Zorbax Eclipse Plus C18 (2.1 mm x 150 mm, 3.5 um) with Water:ACN and 0.1% Formic Acid | 316.8 seconds | 40023050 | | HILIC_BDD_2 = Merck SeQuant ZIC-HILIC with ACN(0.1% formic acid):water(16 mM ammonium formate) | 197.7 seconds | 40023050 | | UniToyama_Atlantis = RP Waters Atlantis T3 (2.1 x 150 mm, 5 um) with ACN:Water and 0.1% Formic Acid | 681.0 seconds | 40023050 | | BDD_C18 = Hypersil Gold 1.9µm C18 with Water:ACN and 0.1% Formic Acid | 183.8 seconds | 40023050 | | UFZ_Phenomenex = Kinetex Core-Shell C18 2.6 um, 3.0 x 100 mm, Phenomenex with Water:MeOH and 0.1% Formic Acid | 885.4 seconds | 40023050 | | SNU_RIKEN_POS = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 207.4 seconds | 40023050 | | RPMMFDA = Waters ACQUITY UPLC BEH C18 with Water:ACN and 0.1% Formic Acid | 235.8 seconds | 40023050 | | MTBLS87 = Merck SeQuant ZIC-pHILIC column with ACN:Water and :ammonium carbonate | 540.7 seconds | 40023050 | | KI_GIAR_zic_HILIC_pH2_7 = Merck SeQuant ZIC-HILIC with ACN:Water and 0.1% FA | 292.7 seconds | 40023050 | | Meister zic-pHILIC pH9.3 = Merck SeQuant ZIC-pHILIC column with ACN:Water 5mM NH4Ac pH9.3 and 5mM ammonium acetate in water | 188.9 seconds | 40023050 |
Predicted Kovats Retention IndicesUnderivatizedDerivatized |
|---|
| GC-MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Experimental GC-MS | GC-MS Spectrum - 1-Pyrroline-2-carboxylic acid GC-MS (1 TMS) | splash10-00di-2900000000-66ac333a416ee8f7a478 | 2014-06-16 | HMDB team, MONA, MassBank | View Spectrum | | Experimental GC-MS | GC-MS Spectrum - 1-Pyrroline-2-carboxylic acid GC-MS (Non-derivatized) | splash10-00di-2900000000-66ac333a416ee8f7a478 | 2017-09-12 | HMDB team, MONA, MassBank | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1-Pyrroline-2-carboxylic acid GC-MS (Non-derivatized) - 70eV, Positive | splash10-00mo-9000000000-a7ad471aae5db0467dfa | 2017-08-28 | Wishart Lab | View Spectrum | | Predicted GC-MS | Predicted GC-MS Spectrum - 1-Pyrroline-2-carboxylic acid GC-MS (Non-derivatized) - 70eV, Positive | Not Available | 2021-10-12 | Wishart Lab | View Spectrum |
MS/MS Spectra| Spectrum Type | Description | Splash Key | Deposition Date | Source | View |
|---|
| Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 10V, Positive-QTOF | splash10-03dj-9700000000-06b3e3d5411a8f705758 | 2017-07-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 20V, Positive-QTOF | splash10-014i-9100000000-8fa6f17fa32d2dc2a376 | 2017-07-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 40V, Positive-QTOF | splash10-0006-9000000000-c48f7e9292b52e3d7a54 | 2017-07-25 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 10V, Negative-QTOF | splash10-03di-5900000000-b4814a13bcbaa249948d | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 20V, Negative-QTOF | splash10-02t9-9400000000-d7552022e5be14c1afea | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 40V, Negative-QTOF | splash10-014l-9000000000-c850159936b776efff87 | 2017-07-26 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 10V, Positive-QTOF | splash10-014i-9300000000-e23eb312ac037d064514 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 20V, Positive-QTOF | splash10-014i-9000000000-4b6f8dab3c747051784c | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 40V, Positive-QTOF | splash10-014i-9000000000-43454e9f111a08bb0994 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 10V, Negative-QTOF | splash10-03di-3900000000-8779645df27f611a2273 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 20V, Negative-QTOF | splash10-014i-9200000000-e49e646efe87cb2ac8f1 | 2021-09-22 | Wishart Lab | View Spectrum | | Predicted LC-MS/MS | Predicted LC-MS/MS Spectrum - 1-Pyrroline-2-carboxylic acid 40V, Negative-QTOF | splash10-00kf-9000000000-b0c27aa3a5a4bfd16ebf | 2021-09-22 | Wishart Lab | View Spectrum |
NMR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted 1D NMR | 13C NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 100 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 200 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 300 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 400 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 500 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 600 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 700 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 800 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 900 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 13C NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum | | Predicted 1D NMR | 1H NMR Spectrum (1D, 1000 MHz, D2O, predicted) | 2021-09-16 | Wishart Lab | View Spectrum |
IR Spectra| Spectrum Type | Description | Deposition Date | Source | View |
|---|
| Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M-H]-) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+H]+) | 2023-02-03 | FELIX lab | View Spectrum | | Predicted IR Spectrum | IR Ion Spectrum (Predicted IRIS Spectrum, Adduct: [M+Na]+) | 2023-02-03 | FELIX lab | View Spectrum |
|
|---|